Sharing analysis code
“An article about computational science in a scientific publication is not the scholarship itself, it is merely advertising of the scholarship. The actual scholarship is the complete software development environment and the complete set of instructions which generated the figures.” - Buckheit & Donoho, 1995 (read the full article here)
This quote nicely summarizes the importance of sharing data, methods and code so that others can evaluate the actual work that was done in a research paper. Luckily, web-based technologies make it very easy to share these materials (and even to share the complete software environment using Docker and Singularity containers). However, just releasing your code without annotation is not very informative because others (and future you!) can't make a lot of sense of it. Two helpful tools to annotate your code are RMarkdown and Jupyter notebooks.
How do tools like R Markdown and Jupyter notebooks make research more reproducible?
- R Markdown files and Jupyter notebooks contain 1) the code that others need to reproduce your work along with 2) the narration that a reader needs to understand your work
- R and Python code can be run within these documents, meaning that in principle others can check whether your work is reproducible
R Markdown
Especially when you do your data analysis in R / RStudio, R Markdown is a very useful tool to put your text and analysis together in one place. It is basically R + Markdown (a markup language to format text). It can be used to write a whole paper, including code to generate figures. This code can be outputted in many formats such as html, pdf and Word. For full documentation see also the R Markdown documentation and this neat cheatsheet (pdf).
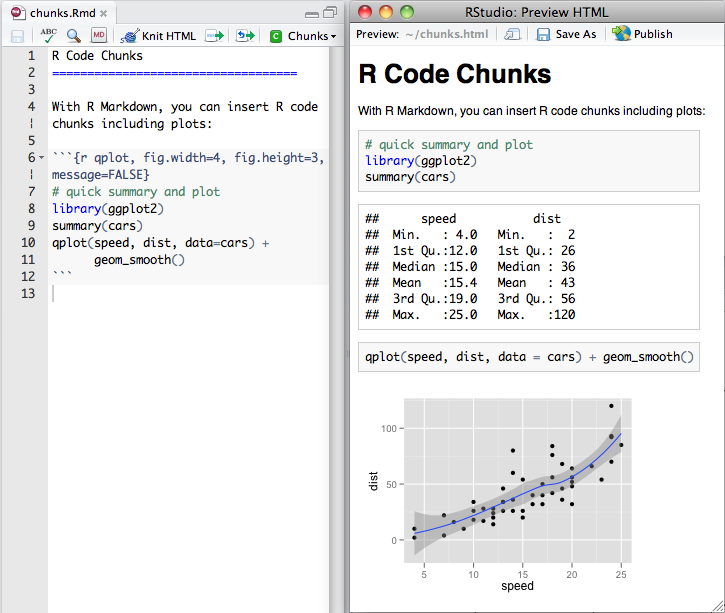
Installing R Markdown
- have R and RStudio installed (avalaible for free)
- install R Markdown
install.packages("rmarkdown")
Reference lists using Zotero in R Markdown
When writing papers, it is also very useful to connect RStudio with Zotero.
Zotero is a free and open source reference manager with a very handy browser plugin. If you have never used a reference manager before: it is a great way to keep a library of all your literature (including pdf's) together and will help you to cite papers in the right way and produce automatic reference lists in the right format for you. This can be done in a word processor like Microsoft Word, but also in R Markdown.
The basic steps you need to make this work:
- install Zotero and import references (e.g., using the browser plugin)
- install the Better BibTex plugin for Zotero by clicking Tools > Add-ons within Zotero and follow these instructions
- install the citr R package
Now when writing text in an RMarkdown file in RStudio:
- within RStudio in the toolbar click Addins > Insert citations
- here you can search through your references and select the ones you want to enter
- you should also edit the YAML header (the upper part of the Rmd file with title, author, output et cetera): here the bibtex file (eg:
bibliography: references.bib) should be listed; - if you also add
# Referencesat the end of the main text, your bibliography will be created automatically there - the format of the bibliography can be defined by pointing to a csl file in the YAML header (e.g,
csl: ./apa.csl). All csl (citation style language) files can be downloaded from the internet, see also https://citationstyles.org/
Jupyter notebooks
For analyses that are conducted using Python, Jupyter notebooks are a great way to keep executable code and annotation in one place (note that many other programming languages are also supported by Jupyter notebooks: the name is reference to the 3 core languages Julia, Python, and R). For full documentation see the Jupyter Notebook docs and https://jupyter.org/ for more information about the larger Project Jupyter ecosystem.
When opening a Jupyter Notebook, you are opening an interactive session. Here you can add different sort of cells: code cells that can be executed (after execution the results will be displayed in the notebook), and Markdown cells that can be used to add descriptive text that can be marked up using the Markdown language.
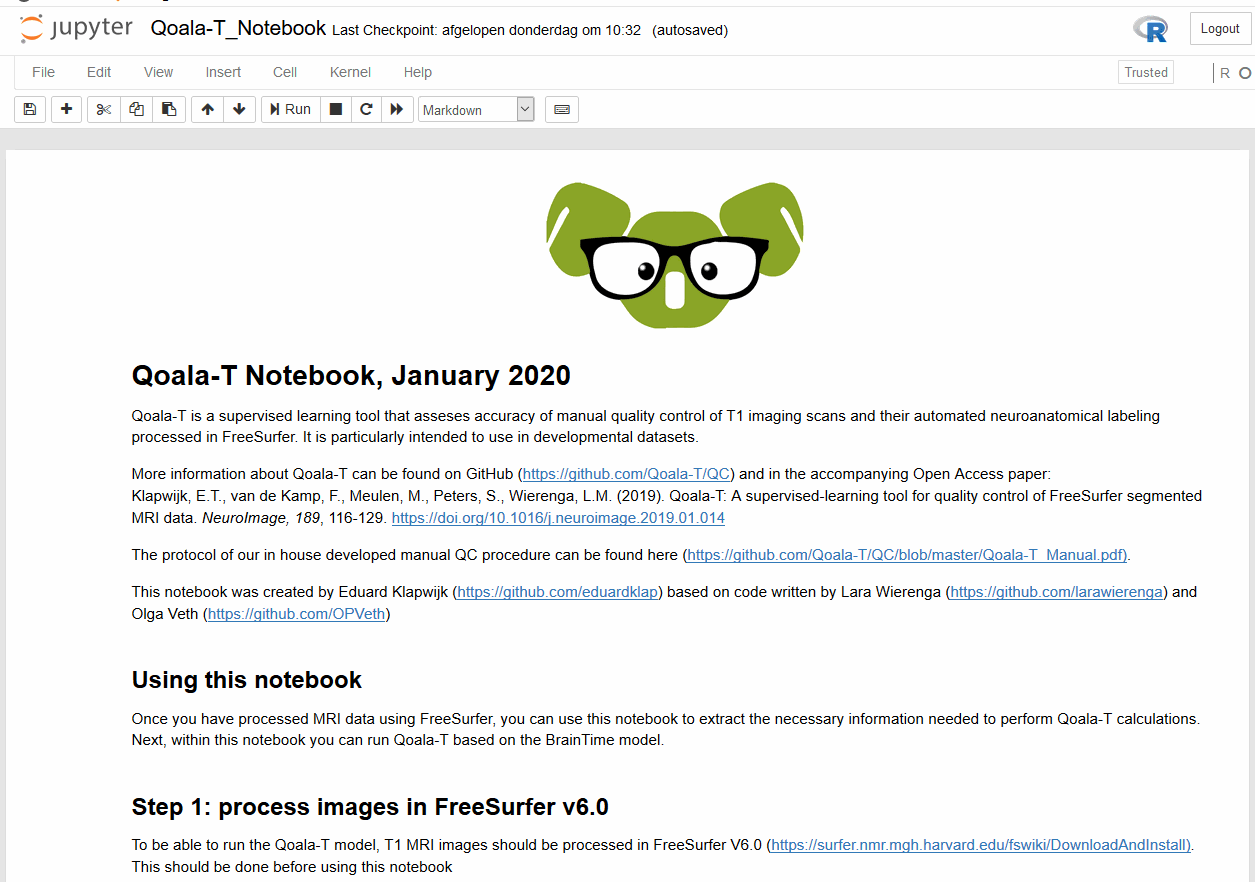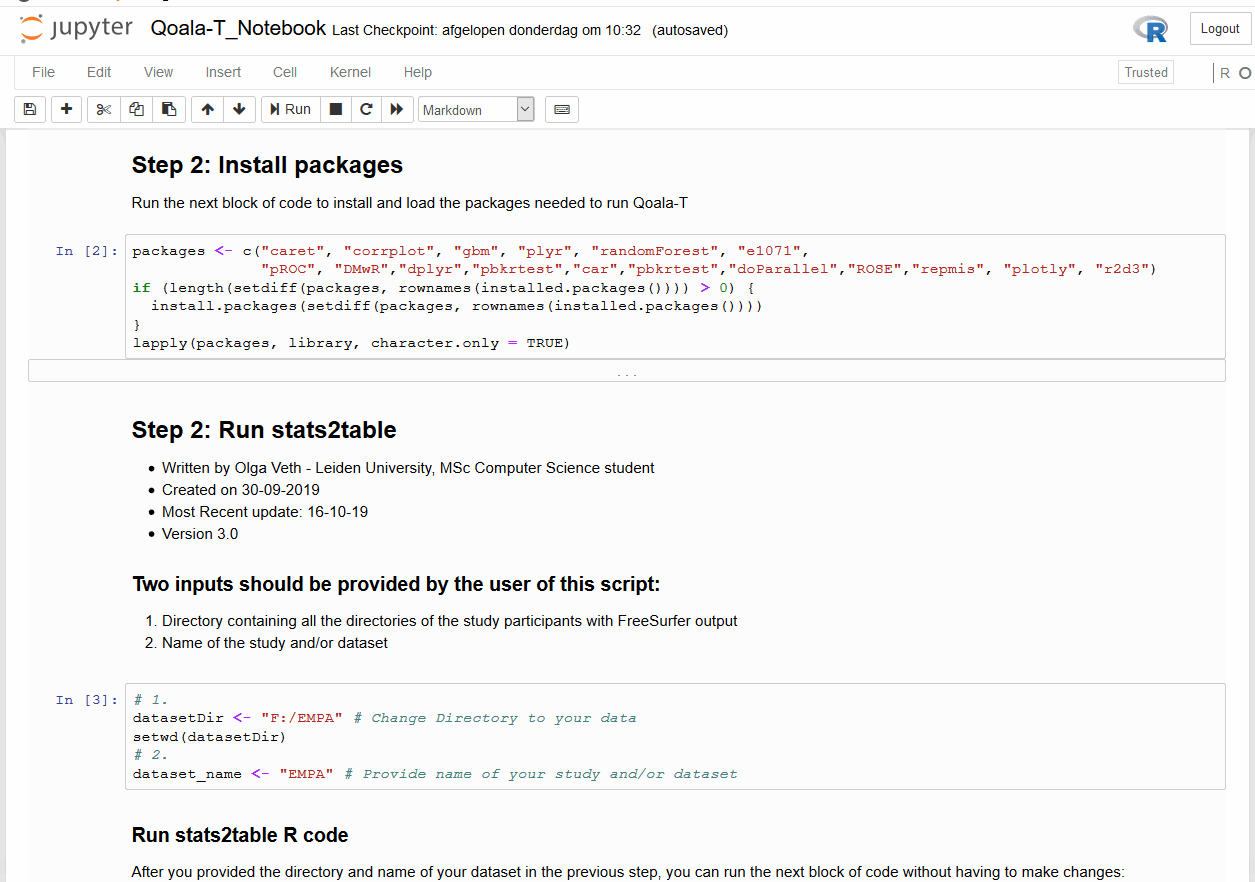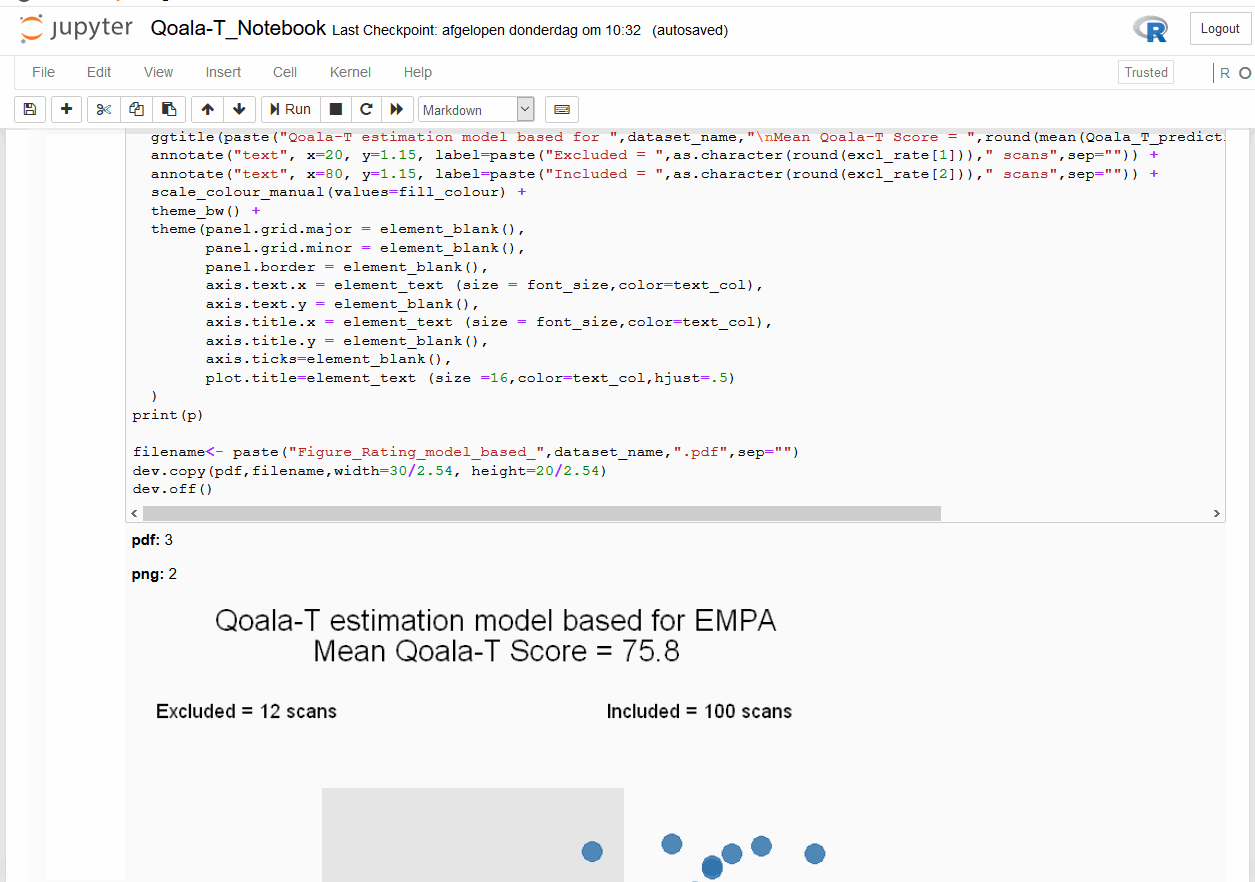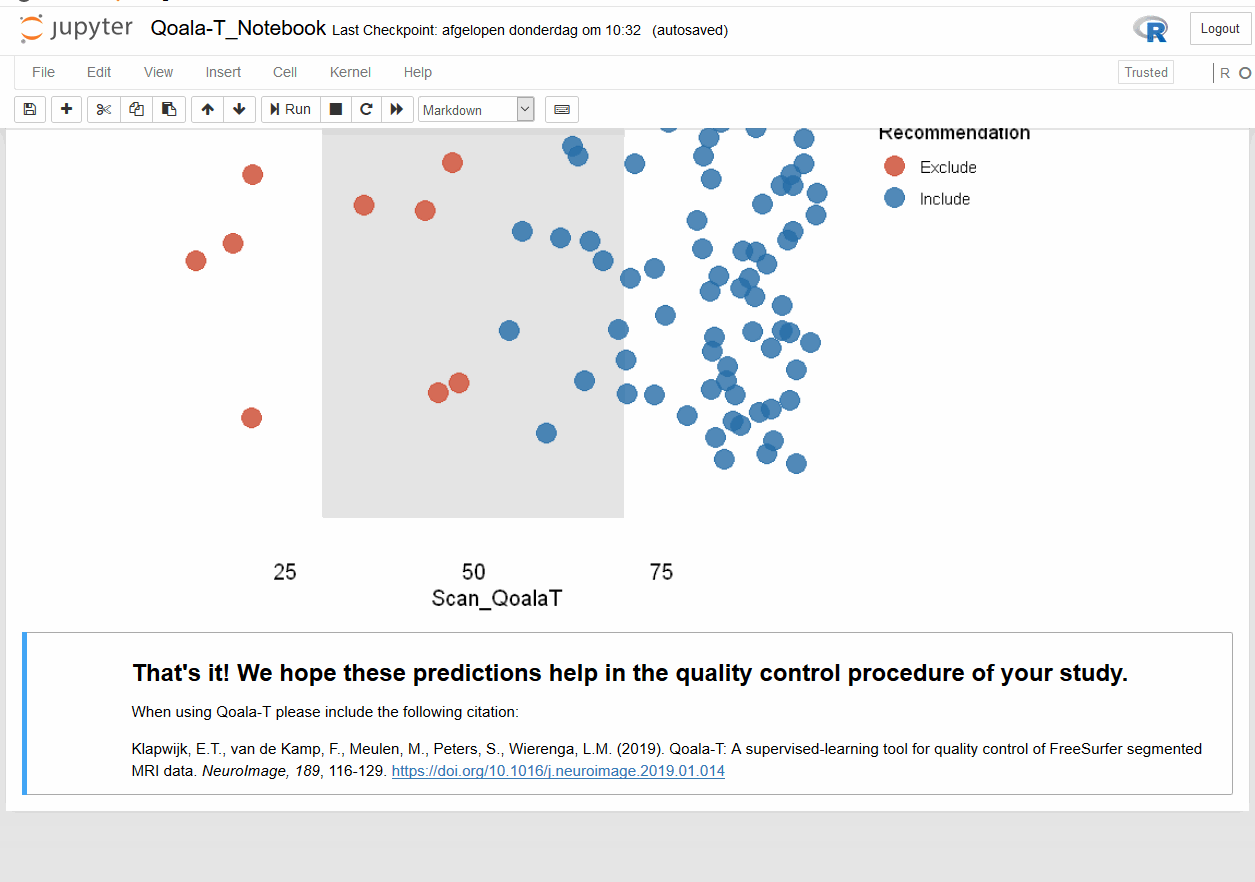
Installing Jupyter notebooks
- you might want to try Jupyter online first, which can be done here
- If you want to install Jupyter notebooks, instructions for the different ways to install it can be found here
- Python is a requirement for installation, and the recommended way to install both Python and Jupyter notebooks is to install Anaconda (available for free)
- Once anaconda is installed you can launch Jupyter Notebooks from the Anaconda Navigator or by typing
jupyter notebookin the command line